![]()
New Work Documents the
Evolution of
Irreducibly Complex Structures
![]()
New Work Documents the
Evolution of
Irreducibly Complex Structures
by Dave Thomas : nmsrdaveATswcp.com (Help fight SPAM! Please replace the AT with an @ . NOTE - ALSO, PLEASE USE A DESCRIPTIVE SUBJECT LINE! E-MAILS WITH NO SUBJECT LINE, OR ONLY A ONE-WORD SUBJECT LINE, WILL BE AUTOMATICALLY DELETED AS SPAM!)
![]()
Introduction
Recent work on the evolution of pigments in star corals, "Evolution of Coral Pigments Recreated," by Juan A. Ugalde, Belinda S. W. Chang, and Mikhail V. Matz, (Science 2004 305: 1433 (9/3/2004), Copyright 2004 AAAS) shows conclusively that "irreducibly complex" structures not only can evolve, but that they have evolved. This should lay to rest the "Intelligent Design" assertion that this type of complexity is forbidden to natural evolution.


The great star coral Montastraea cavernosa, showing cyan-green hues (left) and green-red hues (right) of the coral under fluorescent illumination. Photographs courtesy Mikhail V. Matz (Ph.D.) – Research Assistant Professor , University of Florida. See Copyright Notice below.
![]()
"Irreducible Complexity"
One of the more recent challenges to evolution fostered by the "Intelligent Design" movement involves the concept of "Irreducible Complexity," which was popularized by Michael Behe, a biochemistry professor at Lehigh University, in his 1996 book "Darwin's Black Box." Irreducible Complexity is defined by Behe as follows:
"By irreducibly complex I mean a single system composed of several well-matched, interacting parts that contribute to the basic function, wherein the removal of any one of the parts causes the system to effectively cease functioning. An irreducibly complex system cannot be produced directly (that is, by continuously improving the initial function, which continues to work by the same mechanism) by slight, successive modifications of a precursor system, because any precursor to an irreducibly complex system that is missing a part is by definition nonfunctional. An irreducibly complex biological system, if there is such a thing, would be a powerful challenge to Darwinian evolution. (DBB, p. 39)"
In his presentations, Behe often refers to a few systems as definite exemplars of "Irreducible Complexity," including the bacterial flagellum, the Blood Clotting Cascade, and the Krebs Cycle. Of the Blood Clotting Cascade, Behe remarks
"Blood coagulation is a paradigm of the staggering complexity that underlies even apparently simple bodily processes. Faced with such complexity beneath even simple phenomena, Darwinian theory falls silent. Like some ultimate Rube Goldberg machine, the clotting cascade is a breathtaking balancing act in which a menagerie of biochemicals -- sporting various decorations and rearrangements conferred by modifying enzymes -- bounce off on another at precise angles in a meticulously ordered sequence..." (DBB, page 97)
Behe's point is that if ANY part of the clotting cascade were removed, the system would NOT function at all. He writes
"If the initial action of antithrombin were too fast, the initial action of thrombin too slow, or the original Stuart factor or Christmas factor or antihemophilic factor bound too loosely or too tightly (or if they bound to the inactive forms of their targets as well as the active forms), then the whole system would crash." (DBB, page 95)
Behe says that all "irreducibly complex" structures are barred from evolving because any possible precursor would not have the desired function, and thus could not have been selected for during evolution. Behe is wrong, because he has overlooked the fact that precursor systems may be just fine for functions different from the final, "desired" function.
You can read some critiques of "Irreducible Complexity" arguments at the following websites:
Keith Robison, "Darwin's Black Box: Irreducible Complexity or Irreproducible Irreducibility?"
Ken Miller, "Is the Blood Clotting Cascade "Irreducibly Complex?"
John McDonald, "A reducibly complex mousetrap"
"Behe's Empty Box" by John Catalano
Coral Evolution
How does the new work of Ugalde, Chang and Matz show evolution of "Irreducibly Complex" structures? They studied several different genes of the great star coral Montastraea cavernosa. These various genes code for proteins which generate different fluorescent colors. The colors produced by modern star coral proteins include cyan, shortwave green, longwave green, and red. The authors obtained the sequence information for the modern genes, and performed a phylogenetic analysis with these data. (An example of a simplified calculation of phylogeny from modern protein differences appears in the article "Example Calculation Of Phylogenies: The UPGMA Method.") The Ugalde et. al. analysis produced a detailed history of the key evolutionary events in the divergence of these genes from a common ancestral gene, and produced estimates of the actual genetic and amino-acid sequences of these long-vanished proteins. The authors then genetically engineered these long-extinct proteins into bacteria, grew the bacteria in a culture medium, and then used fluorescent light to observe directly what colors these ancient proteins produced. A stunning visual display of the evolution of these color proteins was developed by using the genetically-engineered microbes to etch the phylogenetic diagram right onto a culture medium, producing a phylogenetic chart in which the color of each branch shows the color of the proteins at that step in the evolution of color genes in the coral. The phylogenetic chart, its fluorescent representation with the engineered bacteria, and the spectra of both modern and ancient proteins appears in Figure 1 of Ugalde et. al., shown below. (This image is copyrighted by Science, and used with permission - please see the Copyright Notice below.) Where there were uncertainties in the amino-acid sequences of ancestral proteins, the authors examined multiple possibilities, and found that none of the sequence ambiguities affected colors to any extent.
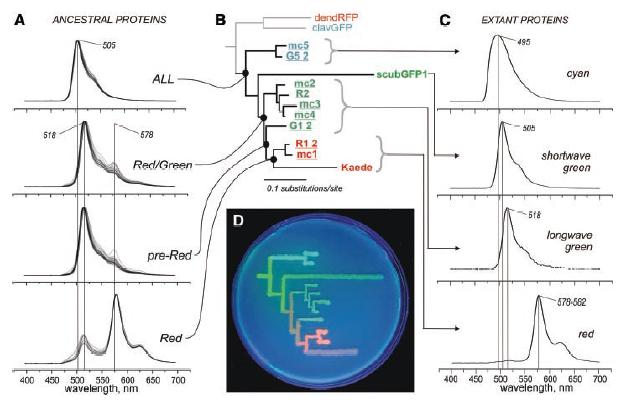
Fig. 1. (A) Fluorescence spectra of the reconstructed ancestral proteins. Multiple curves correspond to clones bearing variations at degenerate sites. (B) Phylogeny of GFP-like proteins from the great star coral M. cavernosa (sequence names are underlined) and closely related coral species. The red and cyan proteins from soft corals (dendRFP and clavGFP) represent an outgroup. (C) Fluorescence spectra of extant proteins. (D) Phylogenetic tree of colors from the great star coral, drawn on a petri dish with bacteria expressing extant and ancestral proteins, under ultraviolet light.
A better-resolution image of Fig. 1 (D) appears below.
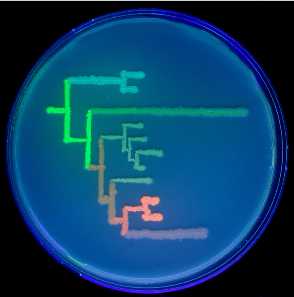
Fig. 1. (D) Phylogenetic tree of colors from the great star coral, drawn on a petri dish with bacteria expressing extant and ancestral proteins, under ultraviolet light.
By actually synthesizing the ancient proteins, the authors were able to explain the evolution of these striking color proteins. In particular, they found that the ancestral protein of all M. cavernosa colors, the "ALL ancestor," was green, with a spectrum peaking at 505 nanometers (nm). Chemical analysis of the green protein shows that it catalyzes two chemical reactions to produce green: a "Stage A cyclization" and a "Stage B dehydrogenation." Simply put, this is an "Irreducibly Complex" system. If either the Stage A cyclization or the Stage B dehydrogenation doesn't occur, there will be NO green color produced! These reactions, along with additional third reactions needed for red hues, appear below, in a figure from Mikhail V. Matz's website at the University of Florida, here labeled Figure 2 (please see the Copyright Notice below).
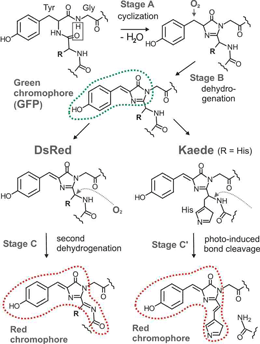
Fig. 2. Chromophore synthesis in green fluorescent proteins (such as GFP) and two convergently evolved red fluorescent proteins DsRed and Kaede (after Cody et al, 1993; Gross et al, 2000 and Mizuno et al, 2003)
Source: http://www.whitney.ufl.edu/research_programs/matz.htm#complexity
Here's where it gets interesting. The coral, like many species, experiences gene duplication events from time to time. Most likely, the ancestral gene for these color proteins was duplicated, leaving the original gene that produced green color, and producing a very similar green-making gene. The new gene was then able to evolve along a different path from its ancestor. One of the evolutionary events in the development of the new gene involved a very small color change, effected by the addition of a third reaction (either a second dehydrogenation, or a photo-induced bond cleavage, see Fig. 2). The effect of this third reaction was minor at first, resulting in the "Red/Green" and "pre-Red" ancestors shown in Fig. 1; these proteins are still primarily green, but have a slight bit of redness (Fig. 1 A). The corals found both colors (green, slighty-reddish green) to be useful, and both were retained. The newer, three-reaction protein wasn't retained because it was more complex; it was retained because corals with that gene survived and were successful. Only after millions of years of subsequent mutations and selection did a form of the gene arise in which the innocuous third reaction ended up catalyzing a brilliant red color. And now, there are several coral colors where once there was but one.
Conclusion
Clearly, if ANY part of the pigmentation cascade were removed from either green-hue or red-hue coral proteins, the systems would NOT function at all. If the "Stage A cyclization" or "Stage B dehydrogenation" were too fast or slow, or otherwise changed, neither green nor red pigments would result. Additionally, if the third reaction (a second dehydrogenation or a photo-induced bond cleavage) was altered in any way, the production of red pigments would collapse.
If the Blood Clotting Cascade is "irreducibly complex," then so too are all of these coral pigment proteins (Green, Cyan, Red, etc.). Each gene codes for proteins that must perform multiple tasks in sequence to achieve the end product (the fluorescent pigmentation).
Behe's claim that any precursor to an "irreducibly complex" system must be non-functional has been disproven. The precursors to the complex red-producing protein didn't produce red, but they were fully functional, producing green with just a hint of red. They provided the "scaffolding" that ultimately led to the complex red protein.
And the evolution of irreducible complexity isn't all that rare, either. Ugalde, Chang and Matz showed that similar red-producing proteins have evolved independently in M. cavernosa. (for example, see dendRFP in Fig. 1B above). Their conclusion:
The more complex red color evolved from green through small incremental transitions (a stepwise accumulation of improvements), each identified in our experiments by ancestral gene reconstruction (Fig 1D). This mode of evolution has been anticipated since Darwin, but has only recently been demonstrated in computer simulation experiments (5, R. E. Lenski, C. Ofria, R. T. Pennock, C. Adami, Nature 423, 139 (2003), "The Evolutionary Origin of Complex Features" ).
"Irreducible Complexity" is no longer an icon of anti-evolution. A major plank of Intelligent Design "theory" has decayed to wood chips and dust. Evolution can produce complex structures, and Ugalde, Chang and Matz have helped to show how and why this happens.
The images on this page are reprinted with permission from Science, and from the authors of "Evolution of Coral Pigments Recreated," by Juan A. Ugalde, Belinda S. W. Chang, and Mikhail V. Matz, which appeared in Science 2004 305: 1433 (9/3/2004), Copyright 2004 AAAS.
Readers may view, browse, and/or download material for temporary copying purposes only, provided these uses are for noncommercial personal purposes. Except as provided by law, this material may not be further reproduced, distributed, transmitted, modified, adapted, performed, displayed, published, or sold in whole or in part, without prior written permission from the publisher.
See the Discussion of this
article on 
For a pleasant diversion, try THOMAS HENRY HUXLEY ON CORAL AND CORAL REEFS
Read More about this work, and about Fluorescent Coral Proteins:
"Glowing Green Proves Darwin Theory" By Jennifer Viegas, Animal Planet News
"BioMedCentral: Color transitions in coral's fluorescent proteins by site-directed mutagenesis"
![]()